Genetic Diversity
ABOUT
The group establishes a bridge between human population and clinical genetics. We study drift, migration, expansion, bottleneck and selection, which model the genetic diversity. This evolutionary framework is applied to identify candidate genes/variants conferring susceptibility to diseases. Our work involves collaborations with anthropologists, statisticians, bioinformaticians and clinicians.
RESEARCH
We are surveying genome-wide chips and whole exome/genome sequences in population samples and in case-control cohorts. These allow unbiased overall evaluations of global human evolution and of candidate genes contributing to complex diseases, respectively. We are particularly interested in investigating how ancestry leads to differential susceptibility to complex diseases. Our current disease models are dengue fever and gastric cancer, and in both African ancestry seems to play a protective role against the worse phenotypes.
We have a long track-record in the study of proteolysis related genes, especially in the context of a common European Mendelian disease, alpha1-antitrypsin deficiency. In addition, we are enlarging our focus in other lung disorders, including Chronic Obstructive Pulmonary Diseases and lung cancer, by applying genomic and proteomic approaches. We are also exploring the involvement of these genes in reproductive biology and in immune response against pathogens, using the seminal hyperviscosity phenotype in male infertility as a model.
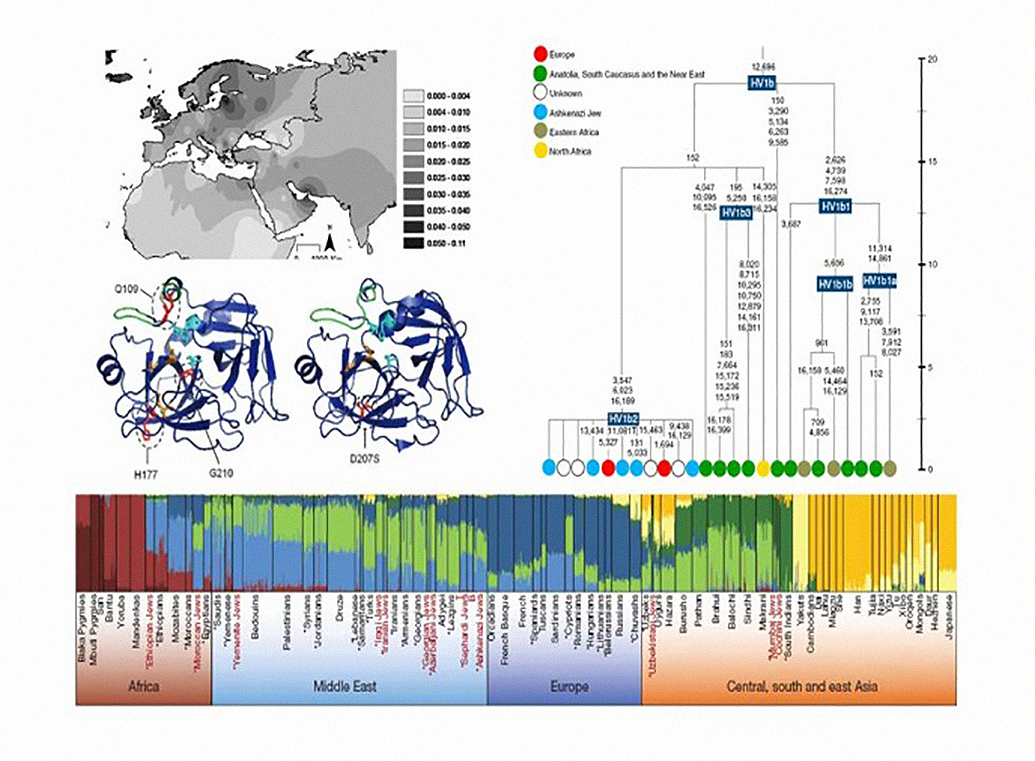
We are internationally recognised by our work in the phylogenetic characterisation of worldwide mitochondrial DNA diversity. As mitochondria play major roles in many life-sustaining functions, they have been implicated in many complex phenotypes, including cancer. We have shown that a proper phylogenetic contextualisation is essential to disentangle between neutral and pathologic variants, and we are currently using this information in researching the cross-talk between the mitochondrial and nuclear genomes.
Team
Selected Publications
Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science356(6337):543-546, 2017. [Journal: Article] [CI: 161] [IF: 41,1]
DOI: 10.1126/science.aal1988 SCOPUS: 85018764496
Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G., Khusnutdinova E.K., Balanovsky O., Semino O., Pereira L., Comas D., Gurwitz D., Bonne-Tamir B., Parfitt T., Hammer M.F., Skorecki K., Villems R.
The genome-wide structure of the Jewish people. Nature466(7303):238-242, 2010. [Journal: Article] [CI: 321] [IF: 36,1]
DOI: 10.1038/nature09103 SCOPUS: 77954542018
Ferreira Z., Seixas S., Andrés A.M., Kretzschmar W.W., Mullikin J.C., Cherukuri P.F., Cruz P., Swanson W.J., Clark A.G., Green E.D., Hurle B.
Reproduction and immunity-driven natural selection in the human WFDC locus. Molecular Biology and Evolution30(4):938-950, 2013. [Journal: Article] [CI: 17] [IF: 14,3]
DOI: 10.1093/molbev/mss329 SCOPUS: 84875635332
Fernandes V., Alshamali F., Alves M., Costa M.D., Pereira J.B., Silva N.M., Cherni L., Harich N., Cerny V., Soares P., Richards M.B., Pereira L.
The Arabian cradle: Mitochondrial relicts of the first steps along the Southern route out of Africa. American Journal of Human Genetics90(2):347-355, 2012. [Journal: Article] [CI: 102] [IF: 11,2]
DOI: 10.1016/j.ajhg.2011.12.010 SCOPUS: 84857046912
Costa M.D., Pereira J.B., Pala M., Fernandes V., Olivieri A., Achilli A., Perego U.A., Rychkov S., Naumova O., Hatina J., Woodward S.R., Eng K.K., MacAulay V., Carr M., Soares P., Pereira L., Richards M.B.
A substantial prehistoric european ancestry amongst ashkenazi maternal lineages. Nature Communications4:, 2013. [Journal: Article] [CI: 71] [IF: 10,7]
DOI: 10.1038/ncomms3543 SCOPUS: 84885452665
Pereira L., Soares P., Radivojac P., Li B., Samuels D.C.
Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. American Journal of Human Genetics88(4):433-439, 2011. [Journal: Article] [CI: 98] [IF: 10,6]
DOI: 10.1016/j.ajhg.2011.03.006 SCOPUS: 79953706954
Soares P., Alshamali F., Pereira J.B., Fernandes V., Silva N.M., Afonso C., Costa M.D., Musilová E., MacAulay V., Richards M.B., Černý V., Pereira L.
The expansion of mtDNA haplogroup L3 within and out of Africa. Molecular Biology and Evolution29(3):915-927, 2012. [Journal: Review] [CI: 202] [IF: 10,4]
DOI: 10.1093/molbev/msr245 SCOPUS: 84857059207
Malik R., Dau T., Gonik M., Sivakumar A., Deredge D.J., Edeleva E.V., Götzfried J., Van Der Laan S.W., Pasterkamp G., Beaufort N., Seixas S., Bevan S., Lincz L.F., Holliday E.G., Burgess A.I., Rannikmäe K., Minnerup J., Kriebel J., Waldenberger M., Müller-Nurasyid M., Lichtner P., Saleheen D., Woo D., Debette S., Maguire J., Cole J.W., Majersik J., Jimenez-Conde J., Lee J.M., Rost N., Pare G., Jern C., Lindgren A.G., Cardenas I.F., Rothwell P.M., Levi C., Attia J., Sudlow C.L.M., Braun D., Markus H.S., Wintrode P.L., Berger K., Jenne D.E., Dichgans M.
Common coding variant in SERPINA1 increases the risk for large artery stroke. Proceedings of the National Academy of Sciences of the United States of America114(14):3613-3618, 2017. [Journal: Article] [CI: 41] [IF: 9,5]
DOI: 10.1073/pnas.1616301114 SCOPUS: 85016951331
Sierra B., Triska P., Soares P., Garcia G., Perez A.B., Aguirre E., Oliveira M., Cavadas B., Regnault B., Alvarez M., Ruiz D., Samuels D.C., Sakuntabhai A., Pereira L., Guzman M.G.
OSBPL10, RXRA and lipid metabolism confer African-ancestry protection against dengue haemorrhagic fever in admixed CUBANS. PLoS Pathogens13(2):, 2017. [Journal: Article] [CI: 57] [IF: 6,2]
DOI: 10.1371/journal.ppat.1006220 SCOPUS: 85014104562
Marques P.I., Fonseca F., Sousa Ť., Santos P., Camilo V., Ferreira Ź., Quesada V., Seixas S.
Adaptive evolution favoring KLK4 downregulation in East Asians. Molecular Biology and Evolution33(1):93-108, 2016. [Journal: Article] [CI: 4] [IF: 6,2]
DOI: 10.1093/molbev/msv199 SCOPUS: 84964766126